Research in the Loudig Lab
Overview
The laboratory has developed technologies to simultaneously extract RNA and DNA from archived specimens, working with tissues that have been stored for more than 45 years. Our analyses demonstrated that nucleic acids stored for extended periods are usable and can be analyzed with high-throughput technologies. Considering the vast amount of human tissues that are archived by hospitals around the United States and the world, as well as the amount of clinical data and follow-up history associated with these specimens, we have successfully demonstrated that retrospective molecular studies can help identify early molecular changes associated with the development of cancer. In recent years, the laboratory has focused its research efforts on the expression analysis of microRNAs, which are small non-coding RNAs that play universal roles in the regulation of gene expression, and whose deregulation has been associated with all the hallmarks of human cancer. Our research efforts have been directed toward the identification of molecular markers that are deregulated in non-invasive stages of cancer, but that play important roles in the transition toward invasive stages. Some of the work that we have done has also been associated with the evaluation of known molecular markers for progression toward distant metastasis. We are also evaluating the detection and identification of circulating biomarkers, using novel technologies and studying circulating exosomes in cancer patients.
Development of molecular assays for recovery and analysis of compromised nucleic acids from formalin-fixed paraffin-embedded tissue specimens
Development of the Extracellular Vesicle Capture by AnTibody of CHoice and Enzymatic Release (EV-CATCHER) assay
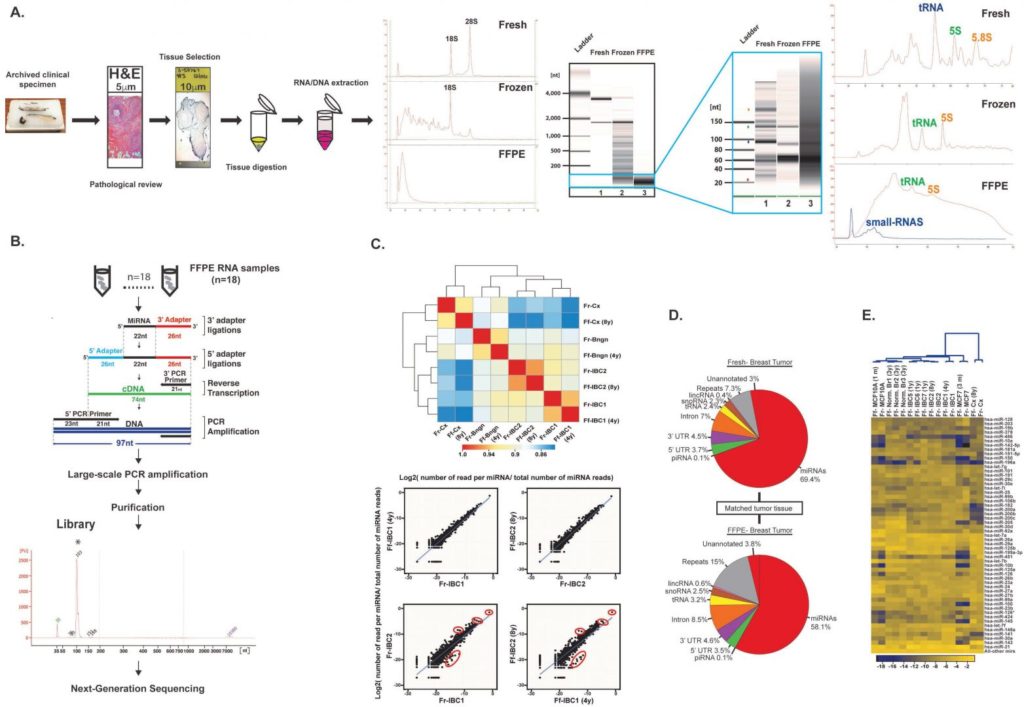
Retrospective microRNA sequencing using archived formalin-fixed paraffin embedded (FFPE) clinical specimens. (A) Tissue processing and pipeline for selection and purification of total RNA from FFPE specimens. Both RNA and DNA are extracted simultaneously. Total FFPE RNA is analyzed on bioanalyzer to evaluate large RNA molecules and small RNAs. (B) Total RNA from FFPE specimens undergoes cDNA library preparation for next-generation sequencing of small RNAs. (C) The miRNA content of matched fresh and FFPE specimens indicates a good correlation between both materials. (D) Distribution of the small RNA content of the matched fresh and FFPE breast tumor specimen evaluated in (C). (E) Clustering analysis of the top differentially expressed miRNAs in normal and tumor breast tissues and cell lines. Ref: Loudig O, et al. Evaluation and Adaptation of a Laboratory-Based cDNA Library Preparation Protocol for Retrospective Sequencing of Archived MicroRNAs from up to 35-Year-Old Clinical FFPE Specimen. Int J Mol Sci. 2017;18(3). pii: E627.
Development of the Extracellular Vesicle Capture by AnTibody of CHoice and Enzymatic Release (EV-CATCHER) assay

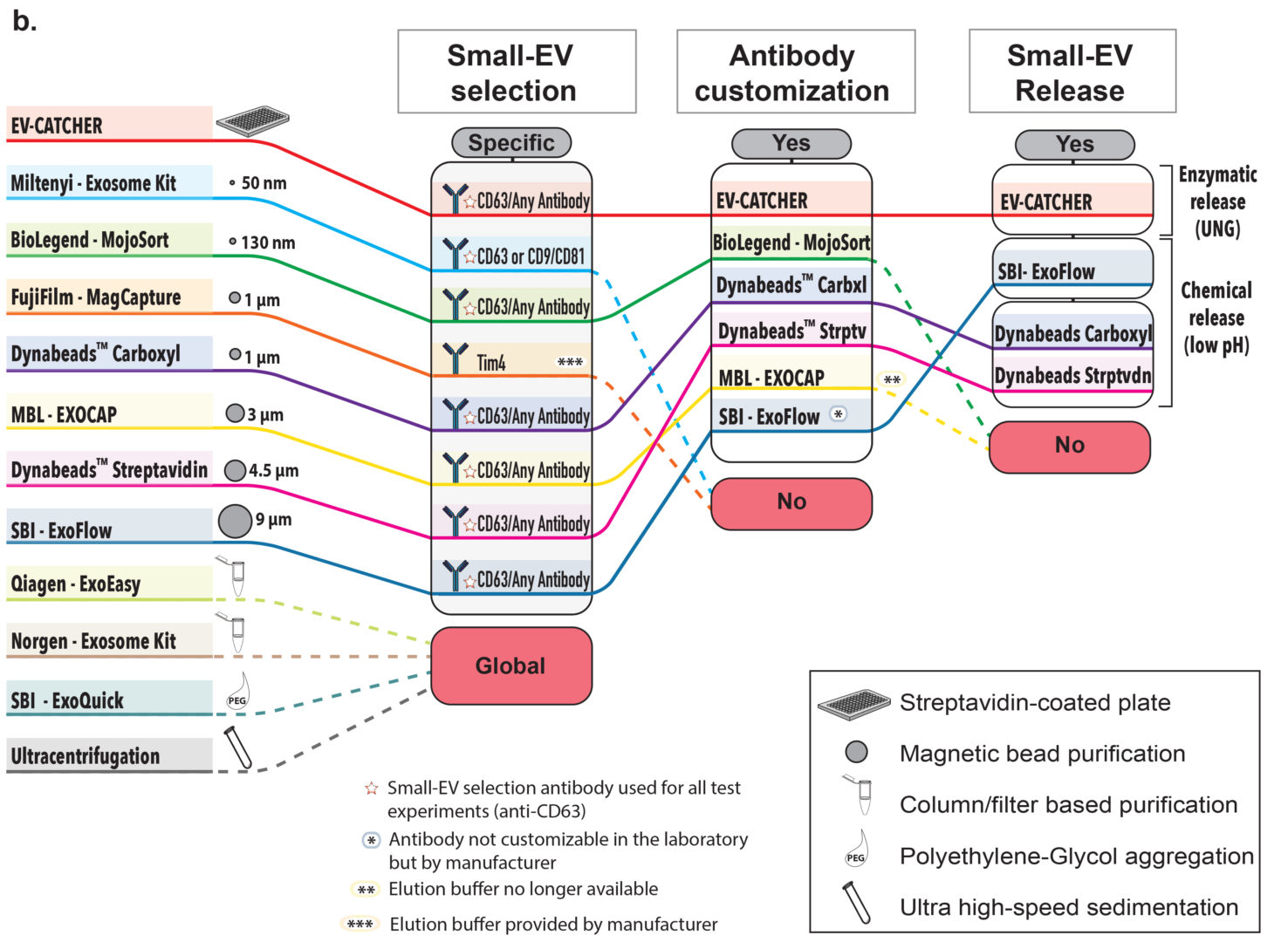
We designed the EV-CATCHER assay to allow for the purification of small-extracellular vesicles (small-EVs) from any biofluid and for any disease. This assay is customizable where a DBCO-activated antibody of choice is initially conjugated to a degradable dsDNA-linker (uracilated 5’-azide oligonucleotide annealed to complementary uracilated 3’-biotin oligonucleotide) and stably anchored to a streptavidin-coated platform (See Figure a., above). The biofluid sample is then incubated overnight for the selective capture of small-EVs. Upon enzymatic treatment with uracil glycosylase (UNG), the selected extracellular vesicles can be released and undergo molecular testing. We compared the EV-CATCHER assay to 11 commercially available and laboratory-based methodologies to evaluate it (See Figure b., above).


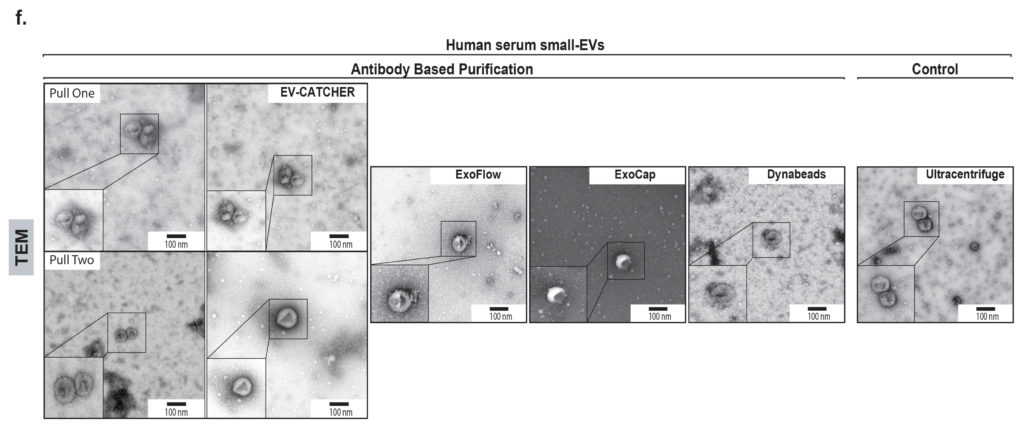
We evaluated the EV-CATCHER assay in comparison with 11 other commercially available and laboratory-based methods, by using ath-miR-159a as a “laboratory-spiked” contaminant in serum samples, prior to the purification of small-EVs. As observed on Figure c (above), the EV-CATCHER assay was superior to all purification methods to prevent the co-purification of small-RNA contaminants. These experiments also revealed that magnetic beads, overall, non-specifically bind small-RNAs. As observed in Figure d., BSA and RNase-A treatments was shown to help decrease the non-specific binding of small-RNAs to magnetic beads, but it remains very high and at levels that can impede the detection of minimal small-RNA expression changes in specific biofluids. When compared to the 11 other methods, EV-CATCHER overall provided clean populations of intact small-EVs (See Figures e. and f.).
EV-CATCHER allows purification of functional SARS-CoV-2 neutralizing small-EVs from the serum of patients infected Covid-19
For these experiments we tested three serum samples obtained from previously infected Covid-19 individuals with high anti-spike IgG titers (High; as defined by >10,000 dilution of sera) and 3 serum samples from individuals with anti-spike IgG levels below limit of quantification (BLQ; not detectable) as shown in Figure a., above . We used our EV-CATCHER assay to purify CD63+ small-EVs from the different serum samples and confirmed their identity by Western blot analyses, which revealed homogeneous purification of small-EVs between high-IgG and BLQ-IgG sera (See Figure b.). We used TEM to assessed the size, morphology, and integrity of the purified EVs from both high and BLQ serum sample (See Figure c.). Small-EV size distribution was also evaluated by nanoparticle tracking (See Figure d.) which revealed similar size distributions (65-150nm) and particle concentrations between high-IgG (average concentration: 1.99 x 1010 particles/ml (n = 20,481)) and BLQ-IgG (1.55 x 1010 particles/ml (n = 20,449)) serum small-EVs. We performed mNeonGreen SARS-CoV-2 reporter virus infection and propagation using gold standard Vero E6, African green monkey kidney cells, which harbor high-levels of the ACE-2 receptor, to evaluate the neutralizing properties of the different sera and our purified circulating serum small EVs. Figure e. displays the purification and dilutions of the serum to obtain small-EVs by ultracentrifugation, which were used as a control to validate the neutralizing properties of circulating small-EVs, purified by EV-CATCHER, from patients recovered from Covid-19 infection. Figure f. displays treatment of VeroE6 cells with whole convalescent sera from individuals with high anti-spike IgG titers that resulted, as anticipated, in neutralization of the SARS-CoV-2 virus (High IgG), but that was not observed when using whole sera from individuals with BLQ IgG titers (BLQ IgG). Our experiments revealed that Vero E6 cells pre-treated with ultracentrifuged small-EVs from a high anti-spike IgG titer serum sample lead to a similar neutralization effect of the SARS-CoV-2 virus (Fig. f, third column) as to that observed with whole sera treatment, whereas cells treated with ultracentrifuged small-EVs from the BLQ anti-spike IgG titer serum sample displayed no neutralizing effect on the virus (Fig. f, fourth column). Based on the extremely high-level of dilution obtained from ultracentrifugation of sera, our data suggest that neutralizing effects observed with ultracentrifuged small-EVs from high anti-spike IgG titer sera were attributable to small-EVs. In order to evaluate the EV-CATCHER assay for in providing intact, functionally neutralizing, and pure antibody-selected small-EVs, we purified small-EVs directly from high anti-spike IgG titer sera (Fig. f. samples 1-3). These small-EVs exhibited a similar neutralizing activity against SARS-CoV-2, as to what was observed solely with ultracentrifuged small-EVs from high anti-spike IgG titer serum but not with BLQ anti-spike IgG sera (Fig. f. samples 4-6). These observations were validated by the detection of fluorescent viral particles by imaging (Fig. g). Altogether these analyses demonstrate that circulating small-EVs purified and released by the EV-CATCHER assay are physically and functionally comparable to those obtained by ultracentrifugation.
Tissues and biopsies archived from human patients from whom the pathological data and clinical history have been recorded represent an invaluable source of information for epidemiological studies and biomarker discovery. However, the high-throughput analysis of degraded mRNA transcripts has been hindered by their small size and low abundancy. To overcome these challenges, we developed a molecular technology to restore damaged mRNA transcripts on a global scale. This methodology is based on the synthesis of short antisense DNA primers, produced from the degraded RNA, which can be used to reverse transcribe complementary sense mRNA transcripts contained in a universal “high-quality” mRNA library. With this method, short and damaged mRNA transcripts can be elongated and analyzed on cDNA microarrays to provide tissue/tumor specific gene expression data (US patent 8,497,067).
A major issue encountered when working with archived clinical specimens resides in their limited availability as well as the impossibility to obtain additional tissue material once the initial block(s) have been exhausted. To address this problem, we designed a highly efficient and robust simultaneous RNA and DNA extraction method, which provides maximum amounts of RNA and DNA without affecting extraction yields for either of them (from US patent 9,309,559). We used this method with human tissues and demonstrated its applicability and efficiency when working with older FFPE tissues. Total RNA extracted with this approach can be analyzed in single or multiplex qPCR assays, by microarrays or bead-arrays, and by next-generation sequencing. Using highly degraded total RNA from FFPE specimens extracted by this method, we also designed a highly reproducible and robust cDNA library protocol for analysis of miRNA expression by next-generation sequencing. Finally, genomic DNA extracted with this method can be subjected to bisulfite treatment and analyzed for methylation patterns, and undergo next-generation sequencing. We are currently developing a next-generation sequencing method to analyze degraded FFPE RNA and determined that the whole-genome DASL assay from Illumina provides reliable quantitative data when using archived specimens.
Loudig O, Milova E, Brandwein-Gensler M, Massimi A, Belbin TJ, Childs G, Singer RH, Rohan T, Prystowsky MB. Molecular restoration of archived transcriptional profiles by Complementary-Template Reverse-Transcription (CT-RT). Nucleic Acids Research Methods. 2007;35(15):e94.
Kotorashvili A, Ramnauth A, Liu C, Lin J, Ye K, Kim R, Hazan R, Rohan T, Fineberg S, Loudig O. Effective DNA/RNA Co-Extraction for Analysis of MicroRNAs, mRNAs, and Genomic DNA from Formalin-Fixed Paraffin-Embedded Specimens. PLoS One. 2012;7(4):e34683.
Loudig O, Wang T, Ye K, Lin J, Wang Y, Ramnauth A, Liu C, Stark A, Chitale D, Greenlee R, Multerer D, Honda S, Daida Y, Spencer Feigelson H, Glass A, Couch FJ, Rohan T, Ben-Dov IZ. Evaluation and Adaptation of a Laboratory-Based cDNA Library Preparation Protocol for Retrospective Sequencing of Archived MicroRNAs from up to 35-Year-Old Clinical FFPE Specimen. Int J Mol Sci. 2017;18(3). pii: E627.
Loudig O, Brandwein-Gensler M, Kim RS, Lin J, Isayeva T, Liu C, Segall JE, Kenny PA, Prystowsky MB. Illumina Whole-Genome DASL Platform: Assessing Its Performance in Formalin-Fixed Paraffin-Embedded Samples and Identifying Invasion Pattern-Related Genes In Oral Squamous Cell Carcinoma. Human Pathology. 2011;42:1911-22.
Identification of molecular markers in benign and non-invasive lesions associated with invasive breast cancer development
Identification of miRNAs differentially expressed in breast carcinoma in situ and correlated with breast cancer development. (A) E-cadherin staining and Localization of lobular carcinoma in situ (LCIS) using Hematoxylin and Eosin staining. (B) Quantitative PCR evaluation of miR-375 during breast lobular neoplasia. (C) MiR-375 in situ hybridization and expression in LCIS cells. (D) MiRNA sequencing using archived DCIS breast tissue specimens from matched DCIS cases (patients who subsequently develop breast cancer) and controls (women who do not develop breast cancer). (E) quantitative PCR validation of miR-375 expression in the same matched DCIS cases and controls. (F) Lentivirus expression of miR-375 in MCF10 cells, which in 3-dimensional culture lose the capacity to form acini and display loss of polarity and dysmoprhic colonies, characteristics on in situ breast carcinoma. (Giricz O, Reynolds PA, Ramnauth A, Liu C, Wang T, Stead L, Childs G, Rohan T Shapiro N, Fineberg S, Kenny PA, Loudig O. Hsa-miR-375 is differentially expressed during lobular neoplasia and promotes loss of mammary acinar polarity. The Journal of Pathology. 2012;226(1):108-19.
Breast cancer is a life-threatening disease and the most common type of cancer in the United States. Early detection is considered to be the first line of defense against this disease and has been responsible for a steep decrease in cancer-related deaths. Breast cancer screening is routinely performed by mammography, supplemental digital breast tomosynthesis, ultrasound, and/or MR imaging, which also allow detection of non-invasive ductal carcinoma in situ (DCIS) lesions. DCIS lesions, which are also called stage 0 breast cancer, represent about 20% of all newly discovered breast cancers. These in situ lesions represent the penultimate stage before invasive disease when following a model of stepwise histological progression from atypia, to hyperplasia, to atypical hyperplasia, to carcinoma in situ, and ultimately to invasive disease. Women diagnosed with DCIS receive aggressive treatment; nevertheless, only 5-14% of them will develop invasive disease within 10 years of their DCIS diagnosis. Although there are known clinical and pathological features as well as protein markers that can help evaluate these lesions, very little is known about the molecular pathways and markers involved in the development of invasive disease. Given the latent interval between DCIS diagnosis and subsequent invasive disease development, as well as the time required for prospectively collecting DCIS specimens, molecular analyses require the use of archived specimens.
Using an anatomically and pathologically related study model within the terminal duct lobular unit, we performed miRNA expression analyses on archived normal lobules, lobular carcinoma in situ, and invasive lobular carcinoma specimens. Our analyses revealed miRNA expression deregulation correlated with the development of invasive disease. One of the deregulated miRNAs, namely miR-375, whose expression was increased in in situ lesions concurrent with invasive breast cancer, validated the molecular relatedness between these lesions and suggested that this miRNA may play an important role in the development of invasive disease. Using the non-tumorigenic MCF-10A cell line, which forms acini in three-dimensional culture and provides an in vitro model for mammary morphogenesis, demonstrated that cells expressing miR-375 were unable to form acini, which lost their cellular polarity. A pilot study on DCIS specimens further validated the upregulation of miR-375 expression in lesions from patients who later went on to develop invasive breast cancer. These findings suggest that early molecular events contribute to the development of invasive disease, which are detectable in archived specimens. By conducting large-scale retrospective studies using archived breast DCIS specimens associated with banked clinical data and patient history, we are further mining for novel molecular markers associated with the development of invasive breast cancer.
Giricz O, Reynolds PA, Ramnauth A, Liu C, Wang T, Stead L, Childs G, Rohan T Shapiro N, Fineberg S, Kenny PA, Loudig O. Hsa-miR-375 is differentially expressed during lobular neoplasia and promotes loss of mammary acinar polarity. The journal of Pathology. 2012;226(1):108-19.
Loudig O, Wang T, Ye K, Lin J, Wang Y, Ramnauth A, Liu C, Stark A, Chitale D, Greenlee R, Multerer D, Honda S, Daida Y, Spencer Feigelson H, Glass A, Couch FJ, Rohan T, Ben-Dov IZ. Evaluation and Adaptation of a Laboratory-Based cDNA Library Preparation Protocol for Retrospective Sequencing of Archived MicroRNAs from up to 35-Year-Old Clinical FFPE Specimen. Int J Mol Sci. 2017;18(3). pii: E627
Arthur R, Wang Y, Ye K, Glass AG, Ginsberg M, Loudig O, Rohan T. Association between lifestyle, menstrual/reproductive history, and histological factors and risk of breast cancer in women biopsied for benign breast disease. Breast Cancer Res Treat. 2017.
Development of molecular assays for precise detection of circulating biomarkers in biofluids
Exosomes have emerged as major intercellular communication vesicles and have also been demonstrated to play critical roles in the development and progression of cancer, notably during metastasis. There is a plethora of lipid bilayer vesicles (different sizes and forms) that are released by cells in the micro-environment and the circulation, and which have been shown to contain proteins and nucleic acids that can have tremendous effects on the recipient cells. We have concentrated our efforts on developing assays to purify exosomes and have been able to robustly sequence the miRNA content of circulating exosomes. Based on preliminary analyses and work that we performed with collaborators, we determined that the method of purification greatly influences the data that will be obtained. Thus, we are currently concentrating our efforts on developing reproducible assays that will allow us to purify selected populations of exosomes, enabling us to identify novel cancer-specific biomarkers. Our exosome research focuses on breast, lung, and prostate cancers.
Ho GY, Jung HJ, Schoen RE, Wang T, Lin J, Williams Z, Weissfeld JL, Park JY, Loudig O, Suh Y. Differential expression of circulating microRNAs according to severity of colorectal neoplasia. Transl. Res. 2015;pii: S1931-5244(15)00071-7.
We thank the National Institutes of Health, the National Cancer Institute, and the Breast Cancer Research Foundation for supporting our current and past research projects.
* Dr. Olivier Loudig is the inventor of EV CATCHER technology licensed to EV Diagnostics, an HMH spin-out company. Dr. Loudig and Hackensack Meridian Health have private equity in this company.
