Research in the Tycko Lab
Our lab has a longstanding research program focusing on genetics and epigenetics in human development and disease. Starting with early studies of genomic imprinting and the function of imprinted genes in pediatric cancers and placental biology, our work has evolved into genome-wide and locus-specific profiling of CpG methylation, both net and allele-specific, for disease gene and functional variant discovery and pathway analysis in autoimmune diseases, neuropsychiatric and neurodegenerative disorders, Down syndrome, and cancers. In this work we have a major focus on cis- and trans-acting effects of local genomic sequences and chromosomal copy number on DNA methylation patterns. We are also studying combinations of epigenetic drugs and immunotherapies in mouse models of cancer.
Trisomy 21 and the trans-acting effects of chromosomal aneuploidies on DNA methylation patterns
Our lab carried out some of the earliest microarray-based studies of gene expression in Down syndrome (DS), with findings on non-linear over-expression of specific genes that remain relevant today. We made useful BAC-transgenic mouse models with physiologically realistic and faithful tissue-specific over-expression of important chromosome 21-linked genes. In our 2010 PLoS Genetics paper, we reported our discovery of recurrent, gene-specific, alterations in DNA methylation in DS cells and tissues—a report that has opened a new field of research on trans-acting genetic-epigenetic interactions, with many papers now from other labs. Based on our more recent comprehensive array-based and methyl-seq study of CpG methylation patterns in DS versus control brain cells and T cells, including chromosomally engineered mouse models from our collaborator Dr. Eugene Yu (Roswell Park Cancer Institute), and our meta-analysis of multiple published studies, we can discern highly recurrent and reproducible trans-acting effects of simple chromosomal aneuploidy on patterns of CpG methylation across the epigenome. We expect that our ongoing studies of this phenomenon will have broad significance for understanding the effects of chromosomal and sub-chromosomal aneuploidies not only in DS, but also in other developmental syndromes and in human cancers.
Li C M, Guo M, Salas M, Schupf N, Silverman W, Zigman W B, Husain S, Warburton D, Thaker H and Tycko B (2006). Cell type-specific over-expression of chromosome 21 genes in fibroblasts and fetal hearts with trisomy 21. BMC Med Genet 7:24. PMID: 16539728
Altered DNA Methylation in Leukocytes with Trisomy 21. Kerkel K, Schupf N, Hatta K, Pang D, Salas M, Kratz A, Minden M, Murty VVS, Zigman WB, Mayeux RP, Jenkins EC, Torkamani A, Schork NJ, Silverman W, Croy BA, Tycko B (2010). Altered DNA Methylation in Leukocytes with Trisomy 21. PLoS Genet 6:e1001212. PMID: 21124956, PMCID: PMC2987931
Xing L, Salas M, Zhang H, Gittler J, Ludwig T, Lin CS, Murty VV, Silverman W, Arancio O, Tycko B (2013). Creation and characterization of BAC-transgenic mice with physiological overexpression of epitope-tagged RCAN1 (DSCR1). Mamm Genome 24:30-43. PMID: 23096997, PMCID: PMC3562396
Mendioroz M, Do C, Jiang X, Liu C, Darbary H, Lang CF, Lin J, Thomas A, Abu-Amero S, Stanier P, Temkin A, Yale A, Liu M-M, Li Y, Salas M, Kerkel K, Capone G, Silverman W, Yu Y.E., Moore G, Wegiel J, Tycko B (2015). Trans-effects of chromosome aneuploidies on DNA methylation patterns in human Down syndrome and mouse models. Genome Biology, 16:263. PMID: 26607552, PubMed Central PMCID: PMC4659173
Do C, Xing Z, Yu YE, Tycko B. (2017) Trans-acting epigenetic effects of chromosomal aneuploidies: lessons from Down syndrome and mouse models. Epigenomics. Feb;9(2):189-207. doi: 10.2217/epi-2016-0138. Epub 2016 Dec 2. Review. PubMed PMID: 27911079; PubMed Central PMCID: PMC5549717.
Yu YE, Xing Z, Do C, Pao A, Lee EJ, Krinsky-McHale S, Silverman W, Schupf N, Tycko B. Genetic and epigenetic pathways in Down syndrome: Insights to the brain and immune system from humans and mouse models. Prog Brain Res. 2020;251:1-28. doi: 10.1016/bs.pbr.2019.09.002. Epub 2019 Oct 24.
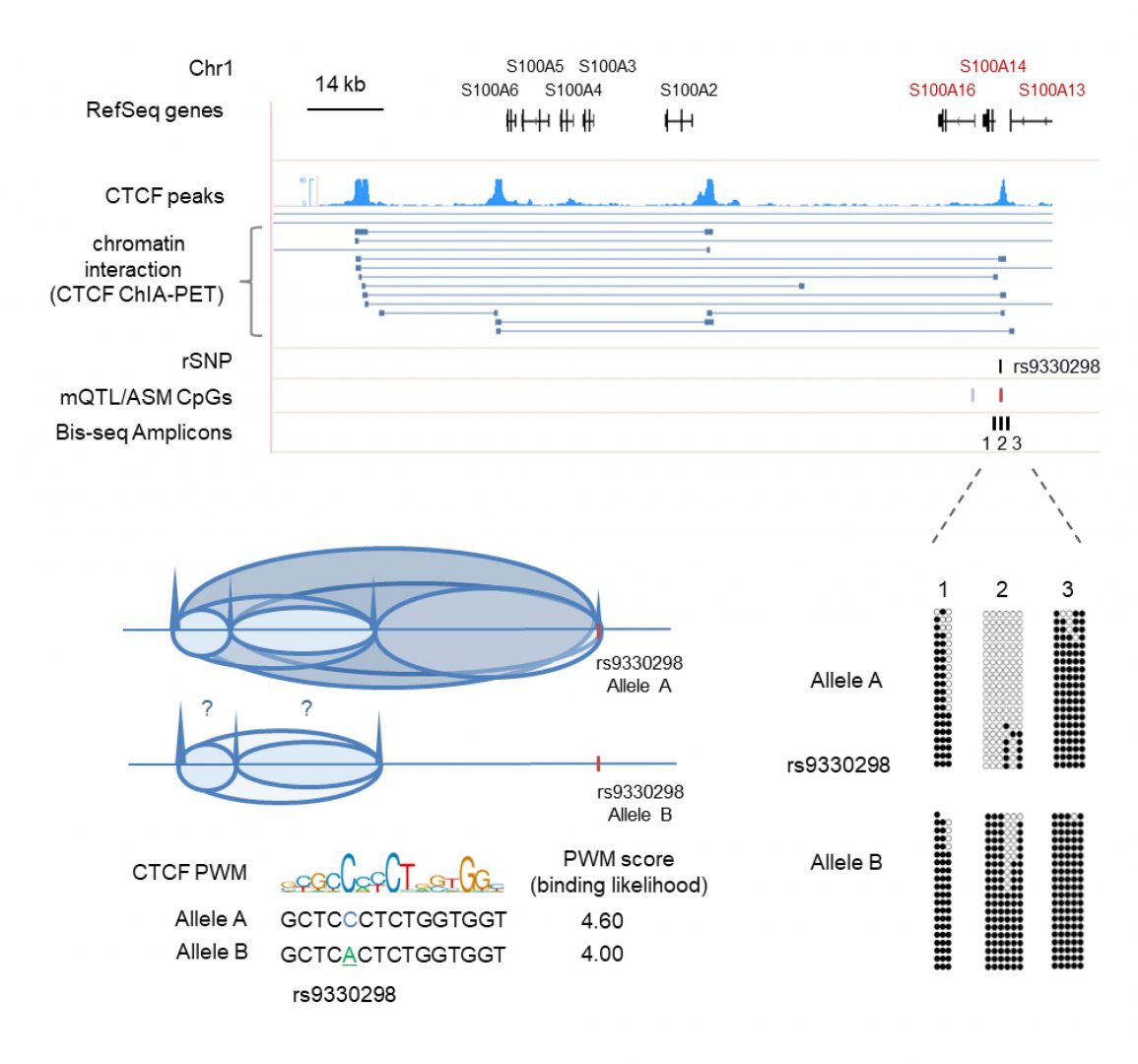
Cis-acting effects of human haplotypes on DNA methylation patterns: basic mechanisms and practical application in post-GWAS mapping
We have a major research emphasis on the mechanisms and consequences of haplotype-dependent allele-specific DNA methylation. This phenomenon, a fundamental cis-interaction between the genome and epigenome, was glimpsed in early studies of VNTR loci in the mid-1980s, but was first uncovered by our lab as a widespread feature of the human epigenome, as reported in our 2008 Nature Genetics paper. In fact, hap-ASM affects more loci than imprinting. In 2010 we proposed that mapping hap-ASM could be a useful practical tool for promoting genome-wide association study (GWAS) peaks to biological true-positives, and more importantly, for honing in on bona fide regulatory sequence variants that underlie supra-threshold GWAS peaks. This idea for “post-GWAS” mapping has since been taken up by many other groups. We are now using array-based methods and Methyl-Seq to generate such maps in primary tissues and overlapping these maps with GWAS data to pinpoint disease-associated regulatory haplotypes. In terms of mechanisms, our 2013 paper in PLoS Genetics raised the interesting possibility of a special involvement of polymorphic CTCF binding sites at insulator elements as underlying some examples of hap-ASM. In our more recent studies (Do, C et al., AJHG 2016, Do, C et al., Genome Biology 2017 and 2020), we validate this mechanism in multiple primary tissues, extend it to polymorphic transcription factor binding sites (TFBS), and test it using cross-species comparisons of TFBS sequences and their associated CpG methylation patterns. Our current emphasis is on using this post-GWAS approach of combined genetic-epigenetic mapping to pinpoint functional genetic variants that underlie susceptibility to cancers, neuropsychiatric, and inflammatory/autoimmune disorders.
Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty V, Schupf N, Vilain E, Morris M, Haghighi F, Tycko B (2008). Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nature Genet 40:904-908. PMID: 18568024
Paliwal A, Temkin AM, Kerkel K, Yale A, Yotova I, Drost N, Lax S, Nhan-Chang CL, Powell C, Borczuk A, Aviv A, Wapner R, Chen X, Nagy PL, Schork N, Do C, Torkamani A, Tycko B (2013) Comparative anatomy of chromosomal domains with imprinted and non-imprinted allele-specific DNA methylation. PLoS Genet 9(8):e1003622. PMID: 24009515, PMCID: PMC3757050.
Do C, Lang CF, Lin J, Darbary H, Krupska I, Velander C, Nagy PL, Petukhova L, Vonsattel JP, Clynes RA, Dwork, AJ, Kral, JG, Monk, C, Christiano, AM, Tycko, B (2016) Mechanisms and disease associations of haplotype-dependent allele specific DNA methylation. Am J of Hum Genet 98: 934-955.
Do C, Shearer A, Suzuki M, Gelernter J, Greally J, Tycko B (2017) Genetic-epigenetic interactions in cis: a major focus in the post-GWAS era. Genome Biology, 18:120.
Do C, Dumont ELP, Salas M, Castano A, Mujahed H, Maldonado L, Singh A, DaSilva-Arnold SC, Bhagat G, Lehman S, Christiano AM, Madhavan S, Nagy PL, Green PHR, Feinman R, Trimble C, Illsley NP, Marder K, Honig L, Monk C, Goy A, Chow K, Goldlust S, Kaptain G, Siegel D, Tycko B (2020) Allele-specific DNA methylation is increased in cancers and its dense mapping in normal plus neoplastic cells increases the yield of disease-associated regulatory SNPs. Genome Biol. Jun 29;21(1):153. doi: 10.1186/s13059-020-02059-3. PMID: 32594908
The tumor microenvironment and anticancer therapies with epigenetic drugs
We were among the first groups to study the role of epigenetics in stromal cells of human gastrointestinal cancers, and we are now studying the efficacy of DNA hypomethylating therapy, both in single-agent and combination therapy protocols, in a mouse model of pancreatic cancer. This line of work is being pursued in close collaboration with our longtime colleague Dr. Tamas Gonda at Columbia University Medical Center.
Jiang L, Gonda T, Gamble M, Salas M, Seshan V, Tu S, Twadell WS, Hegyi P, Lazar G, Steele I, Varro A, Wang TC, Tycko B (2008). Global hypomethylation of genomic DNA in cancer-associated myofibroblasts. Cancer Res 68:9900-9908. PMID: 19047171 PMCID: PMC267054
Quante M, Tu SP, Tomita H, Gonda T, Wang SS, Takashi S, Baik GH, Shibata W, Diprete B, Betz KS, Friedman R, Varro A, Tycko B, Wang TC. (2011) Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell 19(2):257-72. PMID: 21316604, PMCID: PMC3060401
Gonda TA, Kim Y-I, Salas M, Gamble MV, Shibata W, Muthupalani S, Sohn K-J, Abram J, Fox JG, Wang TC, Tycko B (2012). Folic acid increases global DNA methylation and reduces inflammation to prevent Helicobacter-associated gastric cancer in mice. Gastroenterology. 142(4):824-833 PMID: 22474448
Shakya R, Gonda T, Quante M, Salas M, Kim S, Brooks J, Hirsch S, Davies J, Cullo A, Olive K, Wang TC, Szabolcs M, Tycko B#, Ludwig T (2013) Hypomethylating therapy in an aggressive stroma-rich model of pancreatic carcinoma. Cancer Res 73:885-96. PMID: 23204224, PMCID: PMC3548986
Gonda TA, Fang J, Salas M, Do C, Hsu E, Zhukovskaya A, Siegel A, Takahashi R, Lopez-Bujanda ZA, Drake CG, Manji GA, Wang TC, Olive KP, Tycko B. Cancer Res. 2020 Nov A DNA Hypomethylating Drug Alters the Tumor Microenvironment and Improves the Effectiveness of Immune Checkpoint Inhibitors in a Mouse Model of Pancreatic Cancer. 1;80(21):4754-4767. doi: 10.1158/0008-5472.CAN-20-0285. Epub 2020 Aug 14. PMID: 32816859
We thank the National Institutes of Health, the March of Dimes, the Leukemia and Lymphoma Society, the Melanoma Research Alliance, Down syndrome Research Foundation UK, Merck, and Sanofi for supporting our current and past research projects.
